Anti-RPA14 Antibody (56215)
Anti-RPA14 Antibody (56215)
Product No.: 56215
- -
- -
Clone 11.1 Target RPA14 Formats AvailableView All Product Type Monoclonal Alternate Names RP-A p14, Replication factor A protein 3, RF-A protein 3 Isotype Mouse IgG1 Applications IHC FFPE , IP , WB |
Data
![RPA14 antibody [11.1] detects RPA14 protein at nucleus on Mahlarvu xenograft by immunohistochemical analysis. Sample: Paraffin-embedded Mahlarvu xenograft. RPA14 antibody [11.1] (56215) dilution: 1:200. Antigen Retrieval: Trilogy™ (EDTA based, pH 8.0) buffer, 15min](https://www.leinco.com/wp-content/uploads/2025/01/qed-bioscience-anti-rpa14-antibody-56215-1.jpg) RPA14 antibody [11.1] detects RPA14 protein at nucleus on Mahlarvu xenograft by immunohistochemical analysis.
Sample: Paraffin-embedded Mahlarvu xenograft.
RPA14 antibody [11.1] (56215) dilution: 1:200.Antigen Retrieval: Trilogy™ (EDTA based, pH 8.0) buffer, 15min
RPA14 antibody [11.1] detects RPA14 protein at nucleus on Mahlarvu xenograft by immunohistochemical analysis.
Sample: Paraffin-embedded Mahlarvu xenograft.
RPA14 antibody [11.1] (56215) dilution: 1:200.Antigen Retrieval: Trilogy™ (EDTA based, pH 8.0) buffer, 15min - -
- -
Antibody DetailsProduct DetailsReactive Species Human ⋅ Mouse Host Species Mouse Immunogen N-terminally 6x-His tagged fusion protein corresponding to full-length human RPA14 expressed in E. coli. Product Concentration Lot Specific Formulation PBS, pH 7.4 State of Matter Liquid Product Preparation Purified by Protein G affinity chromatography Storage and Handling This antibody is stable for at least one (1) year at -70°C. Avoid multiple freeze- thaw cycles. Regulatory Status Research Use Only Country of Origin USA Shipping Next Day 2-8°C Applications and Recommended Usage? Quality Tested by Leinco Immunoblotting,
Immunoprecipitation: use at 0.2-1 ug/mL. Positive control: HeLa, Raji, and 3T3 cells and recombinant protein. Each investigator should determine their own optimal working dilution for specific applications. See directions on lot specific datasheets, as information may periodically change. DescriptionSpecificity This antibody recognizes human and mouse RPA14. RPA (Replication Protein A) is a single-stranded DNA binding protein. Human RPA is a heterotrimeric protein containing subunits of 70, 32, and 14 kD. This protein complex is highly conserved in eukaryotes and is essential in DNA replication, homologous recombination, and nucleotide excision repair. Function As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism. Thereby, it plays an essential role both in DNA replication and the cellular response to DNA damage (PubMed:9430682). In the cellular response to DNA damage, the RPA complex controls DNA repair and DNA damage checkpoint activation. Through recruitment of ATRIP activates the ATR kinase a master regulator of the DNA damage response (PubMed:24332808). It is required for the recruitment of the DNA double-strand break repair factors RAD51 and RAD52 to chromatin, in response to DNA damage. Also recruits to sites of DNA damage proteins like XPA and XPG that are involved in nucleotide excision repair and is required for this mechanism of DNA repair (PubMed:7697716). Plays also a role in base excision repair (BER), probably through interaction with UNG (PubMed:9765279). Also recruits SMARCAL1/HARP, which is involved in replication fork restart, to sites of DNA damage. May also play a role in telomere maintenance. RPA3 has its own single-stranded DNA-binding activity and may be responsible for polarity of the binding of the complex to DNA (PubMed:19010961). As part of the alternative replication protein A complex, aRPA, binds single-stranded DNA and probably plays a role in DNA repair. Compared to the RPA2-containing, canonical RPA complex, may not support chromosomal DNA replication and cell cycle progression through S-phase. The aRPA may not promote efficient priming by DNA polymerase alpha but could support DNA synthesis by polymerase delta in presence of PCNA and replication factor C (RFC), the dual incision/excision reaction of nucleotide excision repair and RAD51-dependent strand exchange (PubMed:19996105). {PubMed:19010961, PubMed:19116208, PubMed:19996105, PubMed:7697716, PubMed:9430682, PubMed:9765279, PubMed:24332808}. NCBI Gene Bank ID UniProt.org Research Area Cancer Research References & Citations1. Iftode, C et al. 1999. Crit Rev Biochem Mol Biol 34: 141-180 2. Bochkarev, A et al. 1999. EMBO 18: 4498-4504 3. Dong, J et al. 1999. Biochem J 337: 311-317 4. Sibenaller, ZA et al. 1998. Biochemistry 37: 12496-12506 5. Stigger, E et al. 1998. J Biol Chem 273: 9337-9343. Technical ProtocolsCertificate of Analysis |


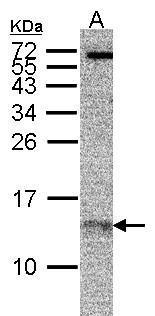
![RPA14 antibody [11.1] detects RPA14 protein by Western blot analysis. A. 30 ug A549 whole cell lysate/extract B. 30 ug MCF-7 whole cell lysate/extract 15 % SDS-PAGE RPA14 antibody [11.1] (56215) dilution: 1:1000](https://www.leinco.com/wp-content/uploads/2025/01/qed-bioscience-anti-rpa14-antibody-56215-2.jpg)